The theme for the third week of Research Data and Digital Scholarship Data Jam 2021 was “Visualizing Text and Networks”.
The “Network Visualization with R” workshop was a live-coding session which detailed the following:
-
Glossary of Network Visualization Terms
-
Software and Programming Alternatives to Network Visualization with igraph and NetworkD3
-
Resources
The R and RStudio demonstration included:
-
Nodes & Edges
-
Directed & Undirected Graph
-
Static and Dynamic Graphs
-
Customizations with layout, colors, & size
Here is the link to the Video Recording of the Workshop.
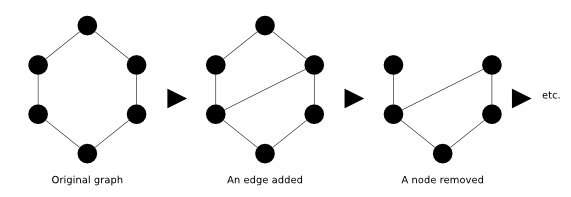
Network visualization allows the viewer to understand a lot of information quickly and intuitively about the state of the network at a glance. Social networks have actors with relations between them. Network diagrams (or Graphs) show interconnections between a set of entities. Each entity is represented by a Node (or Vertice). Connections between nodes are represented by links (or edges).
R Packages commonly used for network visualization include:
- igraph: data preparation and plotting
- ggraph: plotting using the grammar of graphic
- networkD3: interactive graphs
Other relevant R Packages include:
-
Static Network Visualization:
-
igraph
-
ggraph
-
network
-
sna
-
threejs
-
ndtv
-
statnet
-
ggiraph
-
-
Dynamic Network Visualization:
-
networkD3
-
VizNetwork
-
The Python library alternatives are:
-
NetworkX
- igraph
The Julia library alternatives:
-
JuliaGraph
Software alternatives for network visualization are:
-
Gephi
-
Cytoscape
-
Palladio
-
Pajek
For the RStudio demonstration, after installing and running igraph package, a variety of graphs were displayed.
- # Install igraph library
- install.packages("igraph")
- # Run igraph library
- library(igraph)
These included graphs with different number of nodes and edges, directed and undirected graphs, and graphs of different shapes. For example:
- # Create a directed graph with 3 nodes 1, 2, and 3
- g3d <- graph( edges=c(1,2, 2,3, 3, 1), n=3, directed=T)
- plot(g3d)
A simple graph was developed in the form of visualizing a network of historical roads in Philadelphia where the starting point of a street and the ending point of the street are the source and destination, respectively.
- ## Interactive Network Graphs
- ## Historic Streets of Philadelphia
- library(networkD3)
- # Load data
- edges <- read.csv("Historic_Streets.csv")
- head(edges)
- # Identify Source and Target Edges
- source <- edges["FROM_STREET"]
- target <- edges["TO_STREET"]
- # Format into network data
- networkData <-data.frame(source, target)
- simpleNetwork(networkData, nodeColour= "green", zoom=T,
- height = 300, width = 300, fontSize = 16)
The network of interactions in the book Les Misérables by Victor Hugo was also mapped as a force-directed network graph.
- ## Network of Interactions in Les Miserables characters
- # Load data
- data(MisLinks)
- data(MisNodes)
- head(MisNodes)
- head(MisLinks)
- # Plot
- forceNetwork(Links = MisLinks, Nodes = MisNodes,
- Source = "source", Target = "target",
- Value = "value", opacityNoHover = 0.9, NodeID = "name",
- Group = "group", opacity = 0.8, zoom=T)
- # Find all links to Valjean
- which(MisNodes == "Valjean", arr = TRUE)[1] - 1
This graph shows the network of character interactions occurring across the novel at a glance.
Finally, a study of Zachary’s Karate Club Network helped learners understand the use-case of Network Analysis beyond network visualization.
- ## Zachary's Karate Club Network
- library(igraphdata)
- data(karate)
- head(karate)
- # How many factions are in the karate club?
- plot(karate)
- # How many subcommunities clusters are in the karate club?
- cfg <- cluster_fast_greedy(karate)
- plot(cfg, karate)
This graph represents the three major communities and sub-communities within in the network.
It is important to keep in mind the accessibility concerns of network visualizations. For example, overlapping nodes might make the graph seem populated with lower number of nodes. While changing the size and color of nodes might be helpful to discern or distinguish the types of nodes, it might lead to confusion. The 2D graph might be mistaken for occupying 3D space. It is also helpful to use the ColorBrewer2 app to confirm the accessibility features of the colors used in the graph.
Other ways to distinguish nodes, edges, clusters, and networks are as following:
- # nodes/vertices: color, shape, size, border, labels ,text-size, position
- # edges/links: color, size, labels, arrows, curvature, link distance
- # groups/communities: color, shading, border, lines
- # graph: layout, color, faceting
Resources:
-
Static and dynamic network visualization with R, Katya Ognyanova, https://kateto.net/
-
Network Visualization Basics with NetworkX and Plotly in Python, Rebecca Weng, https://towardsdatascience.com/tutorial-network-visualization-basics-with-networkx-and-plotly-and-a-little-nlp-57c9bbb55bb9
-
UPenn LibGuide on Social Network Analysis, Lauris Olsen, https://guides.library.upenn.edu/sna
-
Network Analysis Using R, Yunran Chen, https://yunranchen.github.io/intro-net-r/
-
Temporal Network Visualization Example, http://curleylab.psych.columbia.edu/netviz/netviz5.html#/25
-
Six Degrees of Francis Bacon (Carnegie Mellon), http://www.sixdegreesoffrancisbacon.com/?ids=10000473&min_confidence=60&type=network
-
Awesome Network Analysis Github Masterdoc, https://github.com/briatte/awesome-network-analysis
-
Network R Graph Gallery, https://www.r-graph-gallery.com/network.html
Terminology
-
Brian V. Carolan, “Key Terms,” Social Network Analysis and Education: Theory, Methods & Applications (SAGE, 2014, http://www.sagepub.com/carolan/study/materials/KeyTerms.pdf);
-
Datavu, “Introduction to Network Analysis terminology” (http://datavu.blogspot.com/2013/10/sna-social-network-analysis-basic.html);
-
Katharina Zweig, “An Introductory Course on Network Analysis” (https://sites.google.com/site/networkanalysisacourse/schedule/an-introduction-to-centrality-measures).